Preparing your data for analysis
2025-09-10
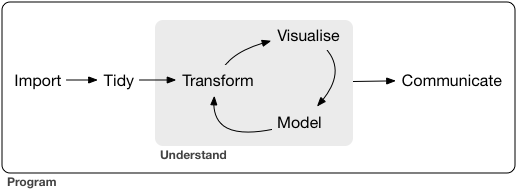
Data science process
Requirements
These notes assume the following…
- you have R and R Studio installed
- at least the
tidyverseandherepackages installed - Converted your Math 615 folder to an R project
- have successfully rendered a quarto document to PDF.
See ASCN Ch 19.1-19.8 for details if you still need help.
Import
- Open your MATH 615 R Project
- Go to Homework 03 and complete step 1 ONLY.
- Confirm your data is in your
datafolder. - Modify the import code on line 13 to correctly import your data set. If you are using one of my data sets, there should be a file in the folder that gives you the correct import code.
- Run this code chunk only (not render)
Confirm import was successful
Okay, did it work?
- Look in the top right Environment pane. Do you see a dataset named
raw? Does it have an expected number of rows and columns? - Click on the table icon to open the data set in a spreadsheet like view. Are the variable names correct? Does the data look correct?
Restrict to analysis variables
- Reduce cogitative load by making a “working” data set that only contains variables that you immediately care about.
Initial Data Screening
Use functions like str() or glimpse() to see what data type R thinks your variables are for the whole data set
Rows: 344
Columns: 3
$ Species <chr> "Adelie Penguin (Pygoscelis adeliae)", "Adelie Penguin (Pygoscelis adeliae)", "Adelie Penguin (Pygoscelis adeliae)", "Adelie Penguin (Pygoscelis adeliae)", "Adelie Penguin (P…
$ Island <chr> "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torgersen", "Torg…
$ `Body Mass (g)` <dbl> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, 4250, 3300, 3700, 3200, 3800, 4400, 3700, 3450, 4500, 3325, 4200, 3400, 3600, 3800, 3950, 3800, 3800, 3550, 3200, 3150, 39…tibble [344 × 3] (S3: tbl_df/tbl/data.frame)
$ Species : chr [1:344] "Adelie Penguin (Pygoscelis adeliae)" "Adelie Penguin (Pygoscelis adeliae)" "Adelie Penguin (Pygoscelis adeliae)" "Adelie Penguin (Pygoscelis adeliae)" ...
$ Island : chr [1:344] "Torgersen" "Torgersen" "Torgersen" "Torgersen" ...
$ Body Mass (g): num [1:344] 3750 3800 3250 NA 3450 ...
- attr(*, "spec")=
.. cols(
.. studyName = col_character(),
.. `Sample Number` = col_double(),
.. Species = col_character(),
.. Region = col_character(),
.. Island = col_character(),
.. Stage = col_character(),
.. `Individual ID` = col_character(),
.. `Clutch Completion` = col_character(),
.. `Date Egg` = col_date(format = ""),
.. `Culmen Length (mm)` = col_double(),
.. `Culmen Depth (mm)` = col_double(),
.. `Flipper Length (mm)` = col_double(),
.. `Body Mass (g)` = col_double(),
.. Sex = col_character(),
.. `Delta 15 N (o/oo)` = col_double(),
.. `Delta 13 C (o/oo)` = col_double(),
.. Comments = col_character()
.. )Both views show you the variable names, data types, and what the data in the first few rows looks like.
Initial Data Screening - single variable
You can also look at the data type for a single variable at a time.
Check plausibility of data values
Confirm these values follow the expected values according to the codebook.
Use table() for categorical variables to see the frequency of unique levels of that variable. Note any levels with less than 10 observations or variables with more than 10 levels.
You can use summary() on numeric variables to see the range of values present. Note the amount of missing values, the spread of the data, and any out of range values (e.g. 99 or 7 on a binary yes/no that should only be 0/1).
Data Prep questions
Questions to ask yourself (and the data) while reviewing the codebook to choose variables to be used in an analysis.
- Are there codes that indicate missing? E.g. MISSING or -99?
- Do you need to make response codes more logical?
- Some systems will record 1=YES and 2=NO. This should be changed to 0=NO.
- Do you need to recode numerical variables to categorical?
- Sometimes categorical data will be recorded as 1, 2, 3 etc when those numbers represent named categories.
- Do you need to create secondary variables such as an average across measures to create a score.
- Are the variable names user friendly? Mixtures of CAPS and lower case, names with spaces or special characters should all be changed. Use the
janitor::clean_names()function to resolve this.
How do I know what needs to be done?
Some of these answers will come only after you look at your data. This can be looking at the raw data itself but also looking at tables and charts generated from the data.
Often when you try to create a plot or table you will encounter an error or something odd looking that will be the notification that something has to be adjusted.
How do I actually make the necessary changes?
- After you identify what you need to do, you have to find an example of how to code the task you want.
- The Applied Stats Course notes has specific examples on how to handle common changes
- Take it one variable at a time.
Looking for help
- The internets can be a wonderful thing when it comes to learning new things.
- The R landscape moves fast. Restrict your search to the past ~4 years.
- Use keywords like
R projector searching for packagevignettes - Stack Overflow and the Posit Community can have gems!
You will start to build up your repertoire of bookmarked sites you like to use.
Using Chat GPT to write code
Example prompt
Using the palmer penguins dataset in R write me R code to change the factor order of the islands variable to put Dream first
- I told it to use a built in data set
- I told it to use R
- I used R terminology such as “factor order” and “variable”
What did it give?
Sure, here’s an example of how you can change the factor order of the island variable in the Palmer Penguins dataset to put “Dream” first using R:
# Load necessary packages
library(palmerpenguins)
# Load the dataset
data("penguins")
# Check the current levels of the 'island' factor
levels(penguins$island)
# Reorder the levels of the 'island' factor to put "Dream" first
penguins$island <- factor(penguins$island, levels = c("Dream", "Torgersen", "Biscoe"))
# Check the new order of levels
levels(penguins$island)In this code, we’re first loading the palmerpenguins package, which provides access to the dataset. Then we load the dataset itself using data("penguins"). We check the current levels of the island factor using levels(penguins$island).
Next, we use the factor() function to reorder the levels of the island variable. We specify the desired order of levels using the levels argument. In this case, we put “Dream” first, followed by the other islands. Finally, we check the new order of levels to make sure the change was successful.
Use it as a learning tool
Not only did it write the code, it wrote literate code. It has code comments (lines 1, 4, 7, prefixed with a #), and a full text explanation.
This is GREAT for learning how to write code!
Plagerism warning
Read the Syllabus on what is expected of you if you use this tool to aid in your writing https://math615.netlify.app/syllabus#use-of-ai
⚠️ Trust but verify! AI is not always correct! Also, this does not replace the necessity of you learning.
Save an analysis-ready data set
- The very last thing you should do in your data management script is save an analysis ready or “clean” data file.
- Advised to save it as a
.Rdatafile to maintain your factor orderings - You may want to do one more round of using
select()to drop variables that you don’t want anymore - Do this at the bottom of your homework dm script file now.
- Every time you render this
dmfile, it will overwrite the cleaned data with the same name - thus ensuring that your analysis data set is always programmatically connected to your raw data set, and all choices documented.
Collaborating with others
- It’s critically important that both of you are using the same slice/filter section of data
- And if one person made a new variable, the other person should have that code also.
- If you and your partner know git – use it.
- Other methods are to store all files (and R project) in a shared Box/Dropbox/Google Drive type folder.
Closing thoughts
- Do not underestimate the importance of this step
- It will take you far, far longer than you anticipate to ‘clean’ your data
- Effort spent here is a direct correlation with payoff.
- Writing code (in any language) will be challenging, but will pay off in the long run
- Don’t reinvent the wheel. If you want to do something, chances are someone else has done it before. Perhaps even yourself!
- Programming in R is not like programming in Python or any flavor of C. Things that are efficient or necessary in those languages (e.g. loops) make your R code unreadable and clunky.